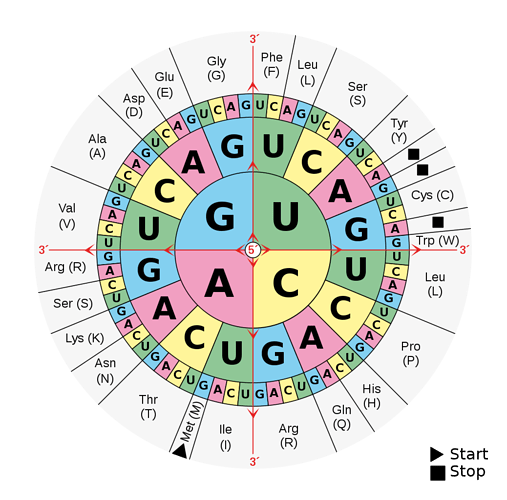
Discussion thread for RNA translation.
The gist of the problem is this:
Given an RNA (string) sequence, translate the three nucleotides into the appropriate aminoacid,
e.g. “UUU” → “Phe”
At first glance, it makes sense to model the table into a directed graph (which would be a tree, since there are no loops). However, there’s another very interesting solution: base 4.
Upon further inspection on the wheel, it’s apparent that there’s a set pattern in the four nucleotide types; UCAG. On every subsection of the wheel, this pattern follows. For anyone experienced with modern-day conventional numeric systems, ex. base 10 which is the one we all use, this will ring a bell, since the same pattern follows (0123456789).
We can extend the same logic, since the nucleotides are accordingly structured.
For example, in base 2 (binary) system, 011 is 3 (in base 10).
In a similar manner, by indexing the wheel starting from UUU, and following in a clockwise rotation until GGG, we can match values 0 to 63 for each combination (64 combinations).
Unsurprisingly, 4^3 (base 4, with 3 “slots”) is, 64.
Which definitely means we can treat the nucleotide combinations as a numeric system without numbers, but nucleotides.
Assume the value of each nucleotide is as such:
U = 0
C = 1
A = 2
G = 3
Then, the combination UUU would translate into 000 (base 4), which unsurprisingly again, translates to decimal 0 (U4^2 + U4^1 + U4^0 → 04^2 + 04^1 + 04^0 = 0).
This matches perfectly with the index we want, which is 0, which in turn matches the aminoacid “Phe”.
Take a moment to think about any combination, and it would match the correct aminoacid!
For the inexperienced, here’s a link that might help further understand: Number Bases (mathisfun)
Equipped with this knowledge, we’re ready to tackle the problem at hand!
First off, we need a way to store the table so we can use it. A simple 64-slot table will do. I’ve created a struct to accomodate for saving the name of the aminoacid, as well as it’s position, which we won’t be using. A valid alternative would be to simply use a char *table[64] instead of the array of structs.
#ifndef SSN184
#define SSN184
#define STOP "STP"
#define START "Met"
#define NECL_BASE 4
#endif
typedef struct aminoacid {
char *name;
int pos;
}Aminoacid;
Aminoacid *create_amino_acid(int index) {
Aminoacid *acid = malloc(sizeof(Aminoacid));
acid->name = malloc(sizeof(char) * 3);
acid->pos = index;
switch (index) {
case 0:
case 1:
acid->name = "Phe";
break;
case 2:
case 3:
acid->name = "Leu";
break;
case 4:
case 5:
case 6:
case 7:
acid->name = "Ser";
break;
case 8:
case 9:
acid->name = "Tyr";
break;
case 10:
case 11:
acid->name = STOP;
break;
case 12:
case 13:
acid->name = "Cys";
break;
case 14:
acid->name = STOP;
break;
case 15:
acid->name = "Trp";
break;
case 16:
case 17:
case 18:
case 19:
acid->name = "Leu";
break;
case 20:
case 21:
case 22:
case 23:
acid->name = "Pro";
break;
case 24:
case 25:
acid->name = "His";
break;
case 26:
case 27:
acid->name = "Gln";
break;
case 28:
case 29:
case 30:
case 31:
acid->name = "Arg";
break;
case 32:
case 33:
case 34:
acid->name = "Ile";
break;
case 35:
acid->name = START;
break;
case 36:
case 37:
case 38:
case 39:
acid->name = "Thr";
break;
case 40:
case 41:
acid->name = "Asn";
break;
case 42:
case 43:
acid->name = "Lys";
break;
case 44:
case 45:
acid->name = "Ser";
break;
case 46:
case 47:
acid->name = "Arg";
break;
case 48:
case 49:
case 50:
case 51:
acid->name = "Val";
break;
case 52:
case 53:
case 54:
case 55:
acid->name = "Ala";
break;
case 56:
case 57:
acid->name = "Asp";
break;
case 58:
case 59:
acid->name = "Glu";
break;
case 60:
case 61:
case 62:
case 63:
acid->name = "Gly";
break;
default:
return NULL;
}
return acid;
}
This is more or less the boilerplate code we need in order to get to the juicy part. It does a few allocations, with a switch/case to match the correct aminoacids. Moving on to the heart of the problem, inside our function we do a few initializations, and we fill the aminoacid table. I’ve made the table static, since we’d only need to initialize the table once every program run. It’s optional, and you may skip both the flag and the static declarations; however you’ll need to free up any heap memory taken up with malloc at the end.
int len = strlen(rna);
int Uv = 0, Cv = 1, Av = 2, Gv = 3;
int index = 0;
static Aminoacid *acid_table[64];
static int run_once_flag = 0;
if (run_once_flag == 0) {
for (int i = 0; i < 64; ++i) {
acid_table[i] = create_amino_acid(i);
}
run_once_flag = 1;
}
And now, for the heart of the problem:
int i = 0 , j = 0, STEP = 3;
while(i < len) {
index = 0;
for (j = i; j < i + STEP; ++j) {
//this should be a hash table for matching, but it's okay
if (rna[j] == 'U')
index += (int) (Uv * pow(NECL_BASE, (STEP - j + i - 1)));
else if (rna[j] == 'C')
index += (int) (Cv * pow(NECL_BASE, (STEP - j + i - 1)));
else if (rna[j] == 'A')
index += (int) (Av * pow(NECL_BASE, (STEP - j + i - 1)));
else if (rna[j] == 'G')
index += (int) (Gv * pow(NECL_BASE, (STEP - j + i - 1)));
}
//Usually, we would return a new array and not modify the RNA, but
// for the purposes of the exercise, we will modify the already given array
//if the name is STP, we stop the process and return everything up until now
if (strcmp(acid_table[index]->name, STOP) == 0) {
memset(&rna[i], '\0', sizeof(char) * (len-i));
break;
}
//always 3 characters
memcpy(&(rna[i]), acid_table[index]->name, sizeof(char) * STEP);
i += STEP;
}
We go over the entire string (rna), with a step of 3 each time for i, in order to scan the ternary patterns.
On every step, we loop over the current position + 3 (for example, on the first while iteration we go from [0, 3) closed open range), and we sum the value to the index. Considering the formula to convert to base 10 with v the value of the number, b the base, and N the number of elements we have this generic formula: vN * b^N + … + v1 * b^1 + v0 * b^0.
Since we’re reading from left to right, and the MSD (Most Significant Digit) is on the leftmost side of the string; and since we’re also steadily reading 3 letters each time, we’re starting off with vN * 4^2 and ending at v0 * 4^0.
Please note that we use this exact formula incrementally, inside the if/else if structures of the for loop.
Afterwards, if the aminoacid is the the same as the defined macro STOP, (UAA, UAG, UGA), we memset the remaining bytes of the array to null (\0) and we stop the entire process.
If the aminoacid is not the STOP signal, we continue on as planned, copying and overwriting the rna array with the names of the aminoacids from the table.
The final program is this (C99):
#include <stdio.h>
#include <stdlib.h>
#include <string.h>
#include <math.h>
#ifndef SSN184
#define SSN184
#define STOP "STP"
#define START "Met"
#define NECL_BASE 4
#endif
typedef struct aminoacid {
char *name;
int pos;
}Aminoacid;
Aminoacid *create_amino_acid(int index) {
Aminoacid *acid = malloc(sizeof(Aminoacid));
acid->name = malloc(sizeof(char) * 3);
acid->pos = index;
switch (index) {
case 0:
case 1:
acid->name = "Phe";
break;
case 2:
case 3:
acid->name = "Leu";
break;
case 4:
case 5:
case 6:
case 7:
acid->name = "Ser";
break;
case 8:
case 9:
acid->name = "Tyr";
break;
case 10:
case 11:
acid->name = STOP;
break;
case 12:
case 13:
acid->name = "Cys";
break;
case 14:
acid->name = STOP;
break;
case 15:
acid->name = "Trp";
break;
case 16:
case 17:
case 18:
case 19:
acid->name = "Leu";
break;
case 20:
case 21:
case 22:
case 23:
acid->name = "Pro";
break;
case 24:
case 25:
acid->name = "His";
break;
case 26:
case 27:
acid->name = "Gln";
break;
case 28:
case 29:
case 30:
case 31:
acid->name = "Arg";
break;
case 32:
case 33:
case 34:
acid->name = "Ile";
break;
case 35:
acid->name = START;
break;
case 36:
case 37:
case 38:
case 39:
acid->name = "Thr";
break;
case 40:
case 41:
acid->name = "Asn";
break;
case 42:
case 43:
acid->name = "Lys";
break;
case 44:
case 45:
acid->name = "Ser";
break;
case 46:
case 47:
acid->name = "Arg";
break;
case 48:
case 49:
case 50:
case 51:
acid->name = "Val";
break;
case 52:
case 53:
case 54:
case 55:
acid->name = "Ala";
break;
case 56:
case 57:
acid->name = "Asp";
break;
case 58:
case 59:
acid->name = "Glu";
break;
case 60:
case 61:
case 62:
case 63:
acid->name = "Gly";
break;
default:
return NULL;
}
return acid;
}
//we're gonna use base 4
char* amino_acid_sequence(char* rna) {
int len = strlen(rna);
int Uv = 0, Cv = 1, Av = 2, Gv = 3;
int index = 0;
static Aminoacid *acid_table[64];
static int run_once_flag = 0;
if (run_once_flag == 0) {
for (int i = 0; i < 64; ++i) {
acid_table[i] = create_amino_acid(i);
}
run_once_flag = 1;
}
int i = 0 , j = 0, STEP = 3;
while(i < len) {
index = 0;
for (j = i; j < i + STEP; ++j) {
//this should be a hash table for matching, but it's okay
if (rna[j] == 'U')
index += (int) (Uv * pow(NECL_BASE, (STEP - j + i - 1)));
else if (rna[j] == 'C')
index += (int) (Cv * pow(NECL_BASE, (STEP - j + i - 1)));
else if (rna[j] == 'A')
index += (int) (Av * pow(NECL_BASE, (STEP - j + i - 1)));
else if (rna[j] == 'G')
index += (int) (Gv * pow(NECL_BASE, (STEP - j + i - 1)));
}
//Usually, we would return a new array and not modify the RNA, but
// for the purposes of the exercise, we will modify the already given array
//if the name is STP, we stop the process and return everything up until now
if (strcmp(acid_table[index]->name, STOP) == 0) {
memset(&rna[i], '\0', sizeof(char) * (len-i));
break;
}
//always 3 characters
memcpy(&(rna[i]), acid_table[index]->name, sizeof(char) * STEP);
i += STEP;
}
return rna;
}